Featured Article
From evolution to molecular biology, scientists use this technology to explore the details
How have the interactions of macromolecules in organisms evolved over eons? To find out, Cuihong Wan and Blake Borgeson—then both working at the Center for Systems and Synthetic Biology at the University of Texas at Austin—used various forms of mass spectrometry (MS) to study protein complexes across a wide range of organisms in search of protein–protein interactions that have endured as organisms evolved. The researchers examined a long list of organisms, from yeast and fungi to mice and humans, and reported: “Clustering reveals a spectrum of conservation, ranging from ancient eukaryotic assemblies that have probably served cellular housekeeping roles for at least one billion years, ancestral complexes that have accrued contemporary components, and rarer metazoan innovations linked to multicellularity.”1
It’s no surprise that Wan et al. used MS to explore the evolution of proteins. “A lot of the applications of MS in the life sciences focus on proteomics,” says Bob Galvin, vice president of ‘omics’ and biopharma at Bruker (Buckinghamshire, U.K.). “Life scientists want to identify as many different peptides—pieces of proteins—as possible.”
Beyond identification, researchers seek to understand the mechanisms that proteins control. So, as Galvin says, “A biologist’s point of view requires more than identifying a protein, but also knowing which ones were up- or down-regulated, which requires accurate quantitation.” A quadrupole time-of-flight (Q-TOF) MS “is very good at quantitation,” says Galvin. The Orbitrap, from Thermo Fisher Scientific (Waltham, Mass.), is well-suited for quantifying peptides, Galvin says.
In addition to gathering quantified information about proteins, life scientists also want to know more about the proteins, such as how they’ve been modified, which is known as post-translational modification. This affects how a protein works and what it does. Adding a sugar to a protein, or glycosylation,2 can make it bind more easily to the outside of cells—think of this as making the protein stickier. This binding can play a role in many crucial processes, including cell proliferation and migration.
Requiring repeatability
 Mass spectrometry can explore the entire spectrum of the life sciences, from simpler organisms—like this green algae—to humans. (Image courtesy of Louisa Howard, Dartmouth College, Hanover, N.H.)
Mass spectrometry can explore the entire spectrum of the life sciences, from simpler organisms—like this green algae—to humans. (Image courtesy of Louisa Howard, Dartmouth College, Hanover, N.H.)“The life science community acknowledges that biological variability is so great that we have to think of new ways to tackle the challenges,” says Mark Cafazzo, director of academic and omics business at SCIEX (Framingham, Mass.). “That typically requires a greater degree of reproducibility and larger study sets than what most proteomics labs are used to.” He adds, “Then you let the data speak for themselves.” And that means sizeable data sets. “The last few years, we have seen growing interest in looking at a large number of proteins in large sample sets to understand differences in populations,” Cafazzo says. “MS offers the ability to dig in and properly identify proteins, which we’ve done for years, and now we can do it with more comprehensive quantitation.”
 Many MS applications in the life sciences involve proteins, including structural analysis. (Image courtesy of George Makhatadze, Rensselaer Polytechnic Institute, Troy, N.Y.)
Many MS applications in the life sciences involve proteins, including structural analysis. (Image courtesy of George Makhatadze, Rensselaer Polytechnic Institute, Troy, N.Y.)SCIEX’s SWATH Acquisition software simultaneously examines thousands of proteins. “If the sample is complex enough,” Cafazzo explains, “SWATH can quantify more than 5000 proteins over a dynamic range of 4.5 linear orders of response.” He adds, “That lets you dig deep into the proteome and see as much as possible, and do so reproducibly.”
In the past, MS identified proteins sequentially, basically one peptide at a time. Today, SWATH is fast enough to collect fragmentation data for all of the detectable ions in a sample in a data-independent approach. “It also does this on a time scale that is appropriate to the liquid-chromatography separation upstream,” Cafazzo explains. “You get a digital archive of all of the fragmentation data for all of the detectable peptides in the sample.”
To generate data even faster, samples can be run at a higher flow rate—say, moving from nanoliters per minute to microliters per minute. To help life scientists manage the data, SCIEX partners with San Diego-based Illumina to provide customers with a data cloud where proteomic data can be combined with genomic data acquired from next-generation sequencing. This approach might go beyond proteomics. “Other life science-oriented disciplines, like lipidomics and metabolomics, could also benefit from such techniques,” Cafazzo says. “You capture more data in less time and throw more data at the problem—letting the statistics overcome the biological variability,” he adds.
Light speed
At the 2015 annual meeting of the American Society of Mass Spec-trometry, Bruker introduced the rapidfleX MALDI Tissuetyper. This matrix-assisted laser desorption ionization (MALDI) imaging platform relies on a high-repetition-rate laser, the smartbeam 3D, which can turn on and off as fast as 10,000 times a second. “With this device, you can actually image tissue samples and look for drug concentration [within] or disease states of tissues directly,” says Galvin. “This links biology with medicine.”
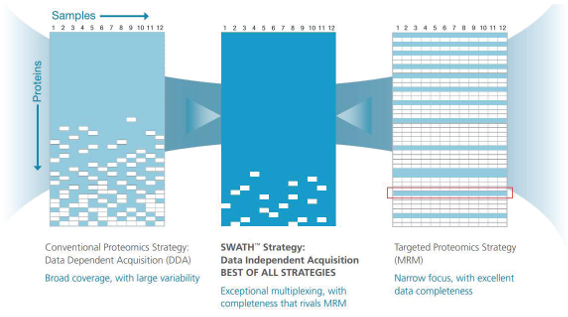 SCIEX’s SWATH Acquisition uses MS to simultaneously examine thousands of proteins. (Image courtesy of SCIEX.)
SCIEX’s SWATH Acquisition uses MS to simultaneously examine thousands of proteins. (Image courtesy of SCIEX.)Getting so much from MS requires advances in hardware, such as a superfast laser, as well as improvements in software. Bruker’s flexImaging software runs the data acquisition and visualization. The platform uses SCiLS Lab software—developed by SCiLS GmbH (Bremen, Germany)—for statistical analysis that can mine data, even with advanced techniques like multivariate analysis.
Today’s MS, says Galvin, turns what once took two days to accomplish into a process that takes just a few hours, and some platforms make that very easy. Bruker’s MALDI Biotyper, which identifies bacteria, “is becoming a black box where you put in a sample, press a button and the instrument does the analysis,” Galvin says. In brief, this platform looks for a unique molecular fingerprint—a collection of proteins—that identifies an organism.
As a result of improving ease of use, more life scientists use MS. According to Galvin, “A decade ago, these instruments were in core facilities, and now you are seeing them in labs where the biologists are doing the analysis directly.” That increases the depth at which life science can be explored, which expands our knowledge of processes behind diseases, evolution and much more.
References
- Wan, C.; Borgeson, B. et al. Panorama of ancient metazoan macromolecular complexes. Nature 2015, 525, 339–44.
- Morelle, W. Michalski, J.-C. Analysis of protein glycosylation by mass spectrometry. Nature Protocols 2007, 2, 1585–1602.
Mike May is a freelance writer and editor living in Ohio. He can be reached at [email protected].