Reproducibility is the bedrock by which scientific validity can be established and one of the central principles underlying scientific practice. However, the reproducibility of a given scientific study can only be demonstrated if the data generated by that study, as well as the methodologies, protocols, and techniques used, are fully disclosed. Reproducibility relies on transparency. Reproducibility cannot be achieved if the data and a detailed description of the methods used to generate them are hidden behind firewalls, be it for technological, economic, legal, or any other reasons.
With laboratories across the world producing millions of data points every day and the biomedical sciences relying on large, multifaceted datasets, reproducibility also requires data to be presented efficiently and in a manner that enables reuse. Datasets are often generated from experiments conducted across multiple laboratories by using diverse methods and standards. Reproducibility relies on these datasets being traceable and integrated by using standard terms and ontologies. At the same time, this also allows researchers to mine data and correlate with other datasets and knowledge.
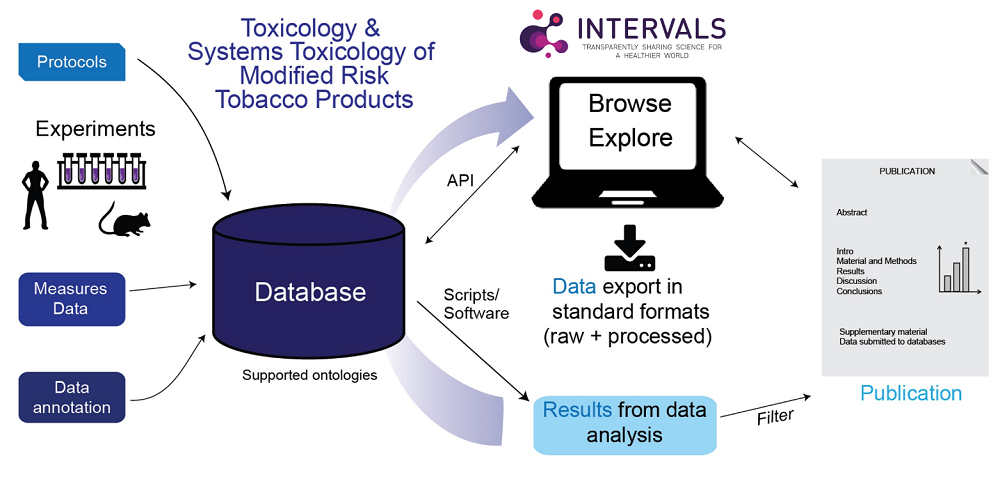
Figure 1 – API: application programming interfaces
Philip Morris International (PMI) launched the research sharing platform INTERVALS to facilitate transparency, data reusability, and reproducibility of results related to the scientific assessment of candidate modified risk tobacco products (cMRTP)*.1,2 Datasets include data from in vivo, in vitro, and clinical studies, generated from traditional toxicology and risk-assessment methodologies as well as from systems toxicology approaches involving -omics measurements and computational assays. Protocols are defined on INTERVALS and directly linked to results and datasets, enabling the community to explore and align best practices with regard to research approaches and methodologies.
The success of INTERVALS, as for any large repository of data that aims to facilitate reproducibility, depends on commitment from the scientific community both to the overarching objectives of effective data sharing and to scientific standards. At the time of writing, most datasets on INTERVALS are from studies conducted by PMI, though parties external to, and independent of, PMI have also started to upload their research. PMI is actively encouraging others to do so. The platform adheres to the FAIR Guiding Principles for data sharing – i.e., data should be Findable, Accessible, Interoperable, and Reusable.2**
Scientific knowledge is generated not only by existing methods accepted by regulators but also by a growing number of alternative research methods and initiatives. To address this, and to ensure the quality of data harmonization, sharing infrastructures need to organize and manage data carefully, allow for processing with a variety of workflows and analysis techniques, and share with the community in a manner that facilitates scrutiny and further analysis. Metadata should be provided that describes experiments, data production, data processing protocols, the use of defined ontologies, and any additional relevant information.
Flexibility of data curation and integration were important considerations in the design of PMI’s platform. New and modified endpoints can be uploaded, while datasets are annotated and verified to allow easy retrieval, interoperability, and meta-analysis. While data and metadata are linked, files are independent so as to ease preparation, possibly by different scientists and laboratories. As such, the datasets are composed of four components: 1) descriptions of metadata formats, 2) metadata files, 3) description of data formats, and 4) data files.
Format descriptions (for both metadata and data) must be uploaded in tabular form and include eight specific columns:
1. Study Stage
This includes all necessary information in five distinct categories: study set-up, treatment, sample preparation, endpoint measurement, and analysis.
2. Column Name
Names of the columns as found in the respective meta(data) files. A relevant ontology term or common vocabulary should be used whenever possible.
3. Role
A set of mandatory (meta)data columns for validation and to allow for analytics and meta-analysis.
4. Type
This specifies the type (e.g., string, float, and int) of (meta)data expected.
5. Ontology
If (meta)data are annotated with ontology terms in the (meta)data files, three pieces of information are required: the human readable term, the ontology the term is taken from, and the identifier of the term. A specific naming scheme for these columns in the corresponding (meta)data files is also expected. Datasets not following the scheme will not pass validation during the upload process.
6. Unit
If values are given in a specific unit, (meta)data files require specification of both the numeric value and unit.
7. Unit Ontology
If the unit is annotated with ontology terms, (meta)data files require specification of the ontology the unit is taken from and the identifier of the unit. This allows for automatic conversion from one unit to another.
8. Description
A free text field with a human-readable description of the (meta)data.
Data files can be divided into tabular and non-tabular datasets. The descriptions of tabular datasets are structured as described above. Non-tabular datasets allow storage of data files in any format. Independent of the tabular/non-tabular categorization, INTERVALS divides datasets into raw, processed, and contrast data files. Raw data files hold sample-specific primary readouts. Processed data files report data generated from this raw data in a processing or analysis step. Contrast datasets report results comparing two different treatments. PMI hopes to compliment the flexible (meta)data storage capabilities of INTERVALS with the possibility to also upload files in standard/regulatory formats (e.g., SDTM, ADAM, SEND, and Isa-Tab).
PMI’s goal is to develop the system to support multi-dimensional data assimilation from sources beyond experimental data generation and publications. While direct quantitative comparisons across studies may not be advisable (given the differences in study design and power), the platform infrastructure allows for a transversal view of results relative to specific diseases and biological pathways. Crucially, by directly linking protocols to results and datasets, the platform facilitates understanding and development of optimal experimental approaches.
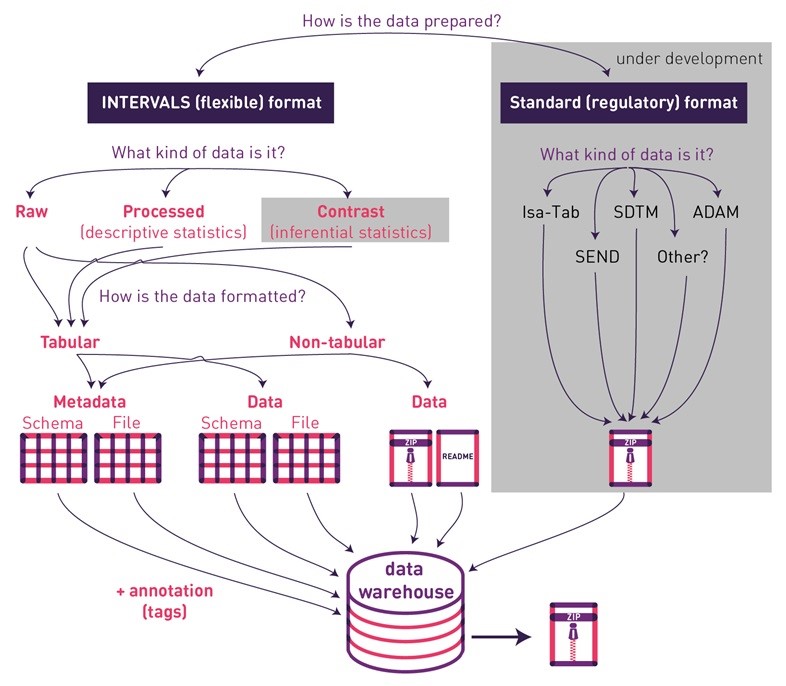
Figure 2 – Dataset upload
The principles and vision for data sharing, integration, and reuse that underly INTERVALS are relevant across the sciences. Other fields that could benefit from this approach include drug discovery and development, medical diagnosis, green chemistry, and many others. PMI’s platform is open to other cMRTP stakeholders to use and contribute to. The broader scientific community are invited to scrutinize the approach with regard to its usefulness for facilitating data integration, reuse, and reproducibility.
*The FDA defines a modified risk tobacco product (MRTP) as a tobacco product that is sold or distributed for use to reduce harm or the risk of tobacco-related disease associated with commercially marketed tobacco products.
**The FAIR Guiding Principles:
Findable
- (Meta)data are assigned a globally unique and persistent identifier
- Data are described with rich metadata (defined in the “reusability” bullet below)
- Metadata clearly and explicitly includes the identifier of the data it describes
- (Meta)data are registered or indexed in a searchable resource
Accessible
- (Meta)data are retrievable by their identifier by using a standardized communications protocol
- The protocol is open, free, and universally implementable
- The protocol allows for an authentication and authorization procedure where necessary
- Metadata is accessible, even when the data are no longer available
Interoperable
- (Meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation
- (Meta)data use vocabularies that follow FAIR principles
- (Meta)data include qualified references to other (meta)data
Reusable
- Meta(data) are richly described with a plurality of accurate and relevant attributes
- (Meta)data are released with a clear and accessible data usage license
- (Meta)data are associated with detailed provenance
- (Meta)data meet domain-relevant community standards
References:
- Intervals. (n.d.). Retrieved from https://www.intervals.science/
- Boué. S. et al. 2017 (last updated: 17 May 2019) Supporting evidence-based analysis for modified risk tobacco products through a toxicology data-sharing infrastructure. F1000Research, 6:12 https://doi.org/10.12688/f1000research.10493.2
- Wilkinson, MD. et al. 2016. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 3:160018 http://dx.doi.org/10.1038/sdata.2016.18