All cells are surrounded by a coating of glycans known as the glycocalyx, and extracellular matrix (ECM) proteins outside of the cell are often glycosylated. Glycosylation is an important process, as it defines intracellular interactions – how cells ‘talk to’ and ‘see’ each other – but it is still an emerging field. For example, glycosylation determines blood groups, and what the body recognizes as ‘self’ and ‘non-self’ in an immunoregulatory capacity. The glycosylation on a cell’s surface is recognized by the immune system and facilitates immune cell binding, particularly in cancer.
In a disease state, changes in glycosylation can be crucial, so research on how glycans change on the surface of cells in response to different disease processes is integral to the discovery and development of new treatments. At the pathological level, changes in glycosylation and their importance in tumor progression have been well documented over time, but until now have not been effectively demonstrated and localization has not been achieved.
Glycan-based biomarkers are currently used for a range of diagnostic purposes, such as in diabetes and cancer. However, most techniques for the identification of these biomarkers are not sufficiently sensitive to be used at early disease stages. Mass spectrometry (MS)-based imaging techniques are breaking new ground in identifying and quantifying the chemical composition of glycans. In contrast with traditional serum-based techniques, MS methods leave the tissue sample intact, and spatial resolution is maintained.
Glycan Imaging
Developments in MS imaging, particularly matrix-assisted laser desorption/ionization (MALDI) Imaging, are enabling researchers to monitor very specific biological changes occurring between different disease states. The powerful spatial localization of MALDI Imaging allows the assignment of different glycans with different tissue features, and these can be translated to glycan changes occurring in blood. Glycan imaging has transformed biomarker discovery, as it can now begin directly in tissue.
Collaborative research at the Department of Cell and Molecular Pharmacology and Experimental Therapeutics, Medical University of South Carolina (MUSC) is driving the development of imaging methods, to better understand disease progression and improve treatments. The Drake, Mehta, and Angel labs collaborate on various aspects of biomarker development, benefitting from sharing expertise and findings.
An advantage of MALDI Imaging for glycan analysis is that their defined masses can be obtained and assessed reproducibly – a crucial feature for translating biomarkers into the clinic. Glycans in formalin-fixed tissues have shown to be excellent targets due to their well-defined structures and the lack of background. The signal is reproducible as it is generated from the activity of the recombinant enzyme peptide N-glycosidase (PNGaseF Prime™) – developed by Dr. Mehta – enabling glycan imaging in tissues for the first time.
For example, in one study the PNGaseF method for MALDI imaging was assessed for archived pathology formalin-fixed paraffin-embedded (FFPE) tissue blocks for liver cancer [1]. An example of liver cancer glycan imaging is shown in Figure 1, as different N-glycans were detected using MALDI quadrupole time-of-flight (QTOF) MS, which could distinguish between non-tumor and tumor regions. Four representative glycan species are shown from a total of 44, as well as an overlay image and segmentation analysis image done in SCiLS Lab software.
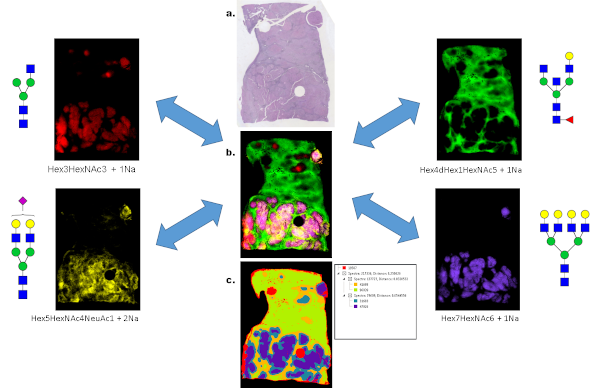
Figure 1: Spatially resolved glycan distributions from PNGaseF digested FFPE human liver cancer tissue. (A) An H&E stain of a hepatocellular carcinoma tissue. The bottom half of the tissue below the red line represents the region of the tumor. (B) A glycan image overlay of four m/z values corresponding to sodium adducts of the N-glycan species shown on the right and left side panels. The major tumor glycan is the Hex7HexNAc6 glycan in blue, and also reported in ref [1]. (C) A segmentation analysis of 5 partitions of 44 N-glycans was done in SCiLS Lab software from Bruker. The distribution patterns of the 5 nodes is shown in the image. Tumor glycans were localized in the blue regions.
Looking Outside the Cell
The composition of the ECM is critical to understanding disease progression, as it contains many proteins and small molecules that contribute to cell communication, and it influences how drugs diffuse between cells during treatment. One of the key challenges of imaging the ECM is that it consists of compact, highly organized ‘bundles’ of proteins which, until now, have been difficult to access analytically. Glycan imaging is an important part of understanding the ECM, as over 90% of ECM proteins are glycosylated. After the Drake lab has studied the glycosylation of a specific tissue, it is passed to the Angel lab to map the changes occurring in the ECM.
The breakthrough development of the PNGaseF enzyme has synergized with Dr. Angel’s research focus on collagen imaging and the glycosylation of ECM glycoproteins. Initially, ECM peptides were being removed from tissue to increase detection of glycans. A strategy was developed to detect the ECM peptides from fixed tissue sections instead of removing them. MALDI Imaging was used to localize collagen and elastin peptides within the tissue microenvironment using bacterial matrix metalloproteinase (MMP) enzyme (Collagenase types; COLases), enabling the team to directly detect localized biochemical information related to ECM collagens and elastin proteoforms [2].
Using this recombinant enzyme allows the group to target ECM proteins in clinical specimens, which had never been done before as there were no tools available to report the changes in these proteins during disease progression. Now, many new interesting changes that are relevant to disease status can be observed.
For example, one application involves examining the ECM of patients with breast cancer, which has a distinct organization through progression and propels the disease in early stages, but is poorly understood (Figure 2). MALDI Imaging can now reveal specific peptides that are modified in the disease state, which could potentially become biomarkers for early stage breast cancer [3].

Figure 2: Example of ECM peptide imaging that shows localized changes in ECM proteins with post-translational modifications of hydroxylated proline (Px) on triple negative breast cancer. Center regions have been collected for tissue microarrays. (A) Serial section Hematoxylin and eosin. Hot pink highlights stromal regions. (B) PTEN tumor suppressor staining. Central region has higher PTEN (brown chromophore). Imaging mass spectrometry was done on the same section that was stained for PTEN. (C) COL3A1 VAVGGLAGYPx m/z 919.4815 (2.1 ppm). (D) COL1A1 GPAGARGNDGATGAA 1242.5825 (-0.24 ppm) E) COL3A1 m/z 1139.6354 GPLGIAGITGARG (-1.8 ppm). Peptides were sequenced and analyzed following Angel et al J Prot Res 2018.
Translating Research to the Clinic
Cancer is an incredibly heterogeneous disease, so multiple biomarkers are needed to detect all cancer types at different stages of progression. The glycan and ECM imaging methods developed by the MUSC group enable this detection and are therefore vital new tools to translate findings into clinical biomarkers. For example, by combining glycopathology markers identified by MALDI Imaging of glycans and ECM proteins in the tumor microenvironment with established histopathology markers, the aggressiveness of a cancer can be established. Glycan and ECM imaging approaches are therefore more than imaging tools, they are emerging as clinically viable tools with significant potential impact in the diagnostic space.
References:
1. West CA, Wang M, Herrera H, Liang H, Black A, Angel PM, Drake RR, Mehta AS (2018) N-Linked Glycan Branching and Fucosylation Are Increased Directly in Hcc Tissue As Determined through in Situ Glycan Imaging, J Proteome Res; 5;17(10):3454-3462.
2. Angel PM, Comte-Walters S, Ball LE, Talbot K, Mehta A, Brockbank KGM, Drake RR. (2018) Mapping Extracellular Matrix Proteins in Formalin-Fixed, Paraffin-Embedded Tissues by MALDI Imaging Mass Spectrometry. J Proteome Res, 17(1):635.
3. Angel PM, Schwamborn K, Comte-Walters S, Clift CL, Ball LE, Mehta AS, Drake RR. (2019) Extracellular Matrix Imaging of Breast Tissue Pathologies by MALDI-Imaging Mass Spectrometry. Proteomics Clin Appl, 13(1):e1700152.
About the Author
Peggi Angel, Ph.D., Assistant Professor, MUSC Proteomics Center, Dept of Cell and Molecular Pharmacology & Experimental Therapeutics, 173 Ashley Avenue BSB 358, Charleston, SC 29425, Tel: (843) 792-8410, Email: [email protected]