by Varouj Amirkhanian, Neo Yang, Eric Tsai, Nicolas Neckelmann, Cecilie Boysen
A simple methodology for detecting viral SARS-CoV-2 RNA from saliva samples using direct reverse-transcription polymerase chain reaction (RT-PCR) and capillary gel electrophoresis with fluorescence detection is presented. The simplified process minimizes the sample collection and treatment steps by eliminating the need for RNA extraction and purification. The treated saliva lysates are directly amplified using a portable thermocycler within 90 minutes, followed by end-point PCR detection utilizing simple-to-use capillary gel electrophoresis instruments in less than 5 minutes per sample run. The Direct RT-PCR Saliva detection platform not only offers a promising option for rapid, accurate and cost-effective COVID-19 testing that avoids the need for costly real time quantitative PCR systems, but it also offers the potential to become portable and eventually available for point-of-care or even at-home use. The detection of other respiratory viruses beyond SARS-CoV-2 in decentralized testing labs is also envisioned.
Nucleic acid detection by real-time quantitative RT-PCR methods is one of the key technologies approved by the China FDA and the US CDC for the diagnosis of COVID-19; however, this technology requires highly trained personnel and can create inaccurate test results (e.g. false negative or positive results). Inaccurate results are caused by inadequate detection sensitivity of quantitative RT-PCR, low viral loads in some patients, difficulties when collecting samples from COVID-19 patients, insufficient sample additions during RT-PCR tests setup, and RNA degradation during the sample handling process. False negative detection can lead to the need to subject patients to multiple tests before a diagnosis can be made; such repeat testing increases the burden on the health care system. Delayed detection and diagnosis can lead to lack of self-isolation of infected patients, or to missing the best time window for treatment.
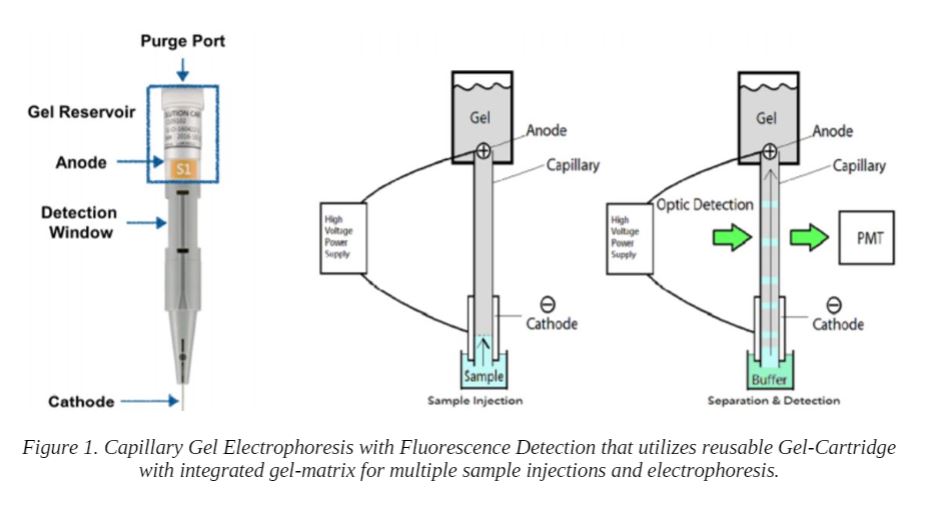
Capillary Gel Electrophoresis with Fluorescence Detection
Capillary gel electrophoresis (CGE), defined as gel electrophoresis carried out in a thin glass capillary in a dedicated instrument, can be used for a diverse range of applications. Automated CGE technology, commonly used in biotechnology laboratories, provides reliable, high resolution and high detection sensitivity at a low cost per sample run. This technology is most often used for the analysis of DNA and RNA, and oligonucleotides, DNA sequencing, as well as for protein and glycoprotein analyses.
We utilized a vertical electrophoresis system that uses reusable gel-cartridges and employs a novel fluorescence detection method (Figure 1). The system uses ball-end optical fibers coupled to an LED for excitation (~ 520 nm) and fluorescence emission (~ 610 nm) is detected directly from the gel cartridge Detection Window.
Portable PCR Thermocycler:
For our experiments we used a portable thermocycler that has been designed to run PCR amplifications of DNA or RNA targets, as well as regular incubations at various temperatures. The small size of the device facilitates the application of molecular biology applications in the field. In practice, the temperature cycles are programmed using a Memory Stick key. Several preprogrammed keys enable the user to quickly set up and switch to other temperature incubations or PCR programs by simply exchanging one pre-programmed application-specific key for another. The portable thermocycler with One-Click-to-Go design, is ideal for basic molecular biology applications used in field laboratories or classrooms covering diverse areas in epidemiology, veterinary sciences, pathogen detection, food testing, ecology, archaeology research and education.
Method and Solution:
To resolve the existing issues associated with real time quantitative RT-PCR, we introduce a novel solution for a highly sensitive RNA detection method that utilizes a compact field portable thermocycler and a Capillary Gel Electrophoresis (CGE) system. The workflow for the analysis of end-point RT-PCR products are shown in Figure 2. Since the CGE system has a much higher fluorescence detection sensitivity than regular gel electrophoresis, it can identify lower copy numbers of viral RNA in the samples collected; SARS-CoV-2 RNA detection is therefore faster and more accurate.

SARS-CoV-2 RNA Virus Detection:
For our detection platform we utilized in-house developed Direct Saliva and RT-PCR reagents that eliminate the need for nucleic acid extraction. This approach provides a unique solution for nontechnical operators in need of rapid reproducible pathogen detection or other genetic testing assays. Endpoint PCR products generated with the RT-PCR reagents are then analyzed with CGE system (Figure 2).For quality control purposes, CGE based detection platform can also be used to analyze PCR products produced by any quantitative real time RT-PCR or regular PCR assays to determine whether the assays are actually producing the expected amplicons. Primer dimer formation and the presence of spurious PCR products can be easily detected.
Detection Process Highlights:
1.Direct RT-PCR amplification. There is no need for RNA extractions and purifications. Saliva samples are mixed with lysis buffer that liberates the nucleic acids. The RNA in these lysates can then be used to set up the RT-PCR amplifications.
2.RT-PCR amplifications are run on the portable thermal cycling system. There is no need for costly real time quantitative PCR instruments.
3.PCR products that have been generated after the full number of cycles (33, 37 or 45 cycles) are then analyzed by CGE utilizing the fully automated Qsep100 Fragment Analyzer with gel cartridges. Results for 1 to 96 samples can be obtained quickly.

Results:
The Qsep100 CGE instrument was used to analyze Direct RT-PCR products in 3 minutes per sample run. Two different kit versions for COVID-19 testing were used to obtain the results presented below (Figure 3 A and B). Gel-image test result demonstrates the detection of RNase P (~292 bp) and the SARS-CoV-2N-gene (~254 bp) in samples derived from COVID-19 Positive and Negative patients (Test Kit Version 1.0 Figure 3A). Note that VAR’s and LaLa’s saliva tested Positive initially and then, when fresh saliva samples were tested 13 or 9 days later, respectively, they tested Negative. Lane 1 corresponds to a Positive Control that verifies that the assay is working as expected. Lane 2 corresponds to the No Template Control (NTC). Figure 3B shows gel-image test results using new primer design and internal controls (Test Kit Version 2.0). The RT-PCR program run on the portable thermal cycler with saliva samples was derived from two patients (KA and KL). The 250 bp amplicon corresponds to the expected size of the target amplicon. KA tested Negative and KL tested positive. A Positive and NTC (No Template Control) were run in the other two lanes (3B).

Summary:
Recent studies of the COVID-19 / SARS-CoV-2 pandemic suggest that frequent testing with a fast turnaround time is what is needed to overcome the current challenges. Surveillance testing may be just as necessary as comprehensive diagnostic testing. We hope that with increased testing, we are better informed about the state of each positive patient and we can reduce the need for lockdowns and protect the most vulnerable populations.
Advantages of the proposed Detection Platform are:
- No RNA Extraction: saving 30 to 60 minutes
- Application Specific thermal cycling: One click to go for 8-samples runs
- Post analysis with CGE system (i.e. Qsep100 Fragment Analyzer) less than 3 minutes per sample
- Multiple Genes analyzed in one reaction: Multiplex Direct RT-PCR
- High Detection Sensitivity: Validation platform for quantitative real time PCR and RT-PCR
- Cost Effective
- Field Portable
Not only does Direct RT-PCR Saliva test offer a promising option for rapid testing, but by avoiding the need for costly real time quantitative RT-PCR, it also has the potential to become portable and eventually accessible for point-of-care or even at-home use. The same strategy can also be expanded to detect and diagnose other respiratory viruses beyond SARS-CoV-2 as well as other pathogens.
About the authors: Varouj Amirkhanian is the Business Development Director at BiOptic, Inc. Meanwhile, Neo Yang is an Application Scientist there, and Eric Tsai is the CEO. Nicolas Neckelmann is an Application Scientist at SNN Altamira Enterprises, and Cecilie Boysen is an Application Scientist at ACBio Consult.
References
1.Meya Kuo, et al. Infectious Diseases Detection Platform: COVID-19. Labcompare, May 15, 2020.
2.Neo Yang, et al. Rapid Genetic Identification of Meat. Labcompare, April 25, 2019.
3.Neo Yang, et al. A Portable CGE System for Cell Free DNA Detection” Labcompare, January 29, 2018
4.DirectSaliva SARS-CoV-2 RNA Virus Detection Kit Instructions. www.bioptic.com.tw