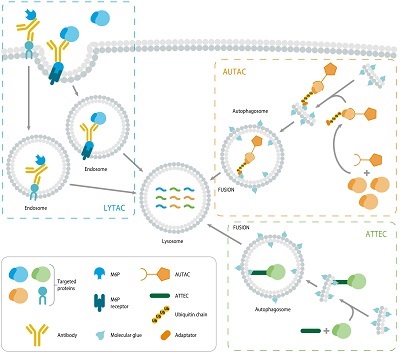
Figure 1: Mechanism of action of degraders based on the autophagy-lysosome system and endosome-lysosome system.
by Fabienne Charrier-Savournin, Life Sciences Technology Platform High Content Reagents Leader, and Volker Eckelt, Director Drug Discovery, Life Sciences Segment, Revvity, Inc.
Abnormal protein expression or activity has been linked to the development of many devastating conditions, including cancer, autoimmune diseases, and neurodegenerative disorders such as Alzheimer’s, Parkinson’s, and Huntington’s. These conditions often involve the misfolding, aggregation, or accumulation of proteins, leading to cellular dysfunction and disease pathology. What if these disease-causing proteins could be selectively eliminated to potentially cure the condition? This is the premise behind targeted protein degradation (TPD), an emerging drug modality that offers the potential to probe biological pathways and target proteins that have previously been considered “undruggable.”
TPD exploits the cellular degradation machinery to selectively target a protein for degradation with the potential for improved therapeutic efficacy compared to traditional small-molecule inhibition and the possibility to discover and validate novel targets. The field of TPD has grown rapidly since it was first described in 20011, with Companies dedicating significant resources to develop TPD-based therapies resulting in over 20 degraders in clinical development2. In addition, there are numerous collaborations between industry and academia focused on advancing the tools and technologies required to develop novel therapeutic degraders.
Endogenous Degradation Systems
The concept behind TPD is to hijack the naturally occurring protein degradation systems within cells using specially designed proteins (“degraders”) that can selectively degrade disease-causing proteins. There are two major endogenous protein degradation systems: the ubiquitin-proteasome system (UPS) and lysosomal degradation pathway (LDP). The UPS is involved in degrading damaged, misfolded, and short-lived proteins, while the LPD is responsible for degrading long-lived proteins and organelles. Both systems are essential for the normal regulation of cellular functions such as cell growth and apoptosis.

Figure 2: Mechanism of action of TRAFTAC degraders.
In the field of TPD, the UPS is the most widely studied of these degradation pathways. Degraders based on this system fall into two main classes: PROteolysis TArgeting Chimeras (PROTACs) and molecular glues. PROTACs are heterobifunctional molecules consisting of two ligands, or “warheads”, connected by a linker. One ligand binds a target protein of interest (POI) while the other recruits and binds an E3 ligase, resulting in the formation of a ternary complex. PROTAC binding induces ubiquitylation of the POI and subsequent degradation by the UPS. By contrast, molecular glues induce or stabilize protein-protein interactions between the POI and an E3 ligase to form a ternary complex. This leads to ubiquitylation of the target and subsequent degradation. Molecular glues do not require a linker and are therefore much smaller in size than PROTAC degraders.
Both PROTACs and molecular glues offer numerous advantages over traditional small-molecule inhibition. For example, they do not need to bind within a biologically active site and do not require tight binding to exert their effects. This means they can be designed to hit previously “undruggable” targets. In addition, recycling of degraders following protein degradation means a single molecule can remove multiple copies of a target protein. They are therefore expected to deliver longer-lasting effects at lower concentrations than small-molecule inhibitors – a property that is particularly attractive for drug development.
While PROTACs and molecular glues have propelled the field of TPD forward, degraders based on the LDP have started to emerge, potentially broadening the scope of degradable targets. The LDP is divided into the autophagy-lysosome system and endosome-lysosome system and degraders based on these systems include AUTOphagy-TArgeting Chimera (AUTACs), LYsosome-TArgeting Chimeras (LYTACs) and autophagosome-tethering compounds (ATTECs) (Figure 1).
AUTACs target a POI for degradation through the autophagy pathway, which is responsible for the degradation and recycling of proteins, organelles, and other cytoplasmic material. AUTACs typically consist of two warheads, one that binds the POI and another that induces autophagy, joined by a linker. Upon binding, the ternary complex redirects the POI to the autophagosome for subsequent lysosomal degradation. Similar to AUTACs, LYTACs also consist of three components, including a ligand for the POI, a linker, and a lysosome-targeting warhead. Upon ternary complex formation, the LYTAC facilitates the transport of the protein to the lysosome via vesicular trafficking pathways, where the POI is degraded. LYTACs typically degrade membrane or extracellular proteins. ATTECs are monovalent degraders that directly link a POI warhead to a ligand. ATTECs act by directing a POI to the autophagosome for subsequent lysosomal degradation.
In addition to PROTACs, AUTACs, and LYTACs, numerous other “TACnologies” are under investigation. These include TRAnscription Factor-TArgeting Chimeras (TRAFTACs) that target transcription factors (Figure 2), PHOtochemically TArgeting Chimeras (PHOTACs) that utilize light-activatable groups to induce protein degradation, and folate-PROTACs that are designed to induce degradation of a POI in cells with high expression of folate receptors. CLIPTACs (in-cell CLIck-formed Proteolysis-TArgeting Chimeras) rely on two individual small precursor molecules that self-assemble to form a functional PROTAC in cells. With lower molecular weights than PROTACs, CLIPTACs are expected to demonstrate enhanced cellular permeability. The use of PHOspho-dependent PROTACs (PHOTACs) has also been reported. This is a conditional degradation method which depends on protein phosphorylation status and is believed to efficiently impair protein functions. Another approach is the development of degrader-antibody conjugates (DACs), which are similar to antibody-drug conjugates, but the cytotoxic payload is replaced with a PROTAC or molecular glue degrader. All these advancements are described in detail by Yang et al. elsewhere3.
Emerging Technologies to Enhance Degrader Discovery and Optimization
While there are a broad range of biochemical and biophysical techniques to study TPD in vitro, the available methodologies for investigating degraders in cellular or animal models are limited. Challenges arise due to the complex nature of intracellular environments and the need to evaluate the effects of degraders within these contexts. One of the key considerations for degrader development is establishing workflows that extensively scrutinize every step of the degradation pathway. Degrader hit identification and optimization is complex and challenging, necessitating accurate assessment of ternary complex formation, target ubiquitination, and subsequent degradation. While the TPD field has long used Western blots as the traditional method for measuring the activity of degradation-activating compounds, high-throughput approaches that quantitatively assess the efficiency, sensitivity, and reproducibility of TPD are needed to guide drug discovery efforts.
The Cellular Thermal Shift Assay (CETSA) is a convenient high-throughput screening (HTS)-compatible approach to assess the capability of degraders to bind to a targeted protein in a cellular context. This method, which can determine target engagement, allows both compound permeability and binding across different cell types to be monitored without any modifications.
Figure 3: HTRF (Homogeneous Time-Resolved Fluorescence) and Alpha (Amplified Luminescent Proximity Homogeneous Assay) no-wash immunoassays can be used to monitor the impact of degraders on a POI over time, in their cellular context, and without the need for molecular engineering.
Permeable compounds’ induced protein degradation can be followed with the luminescent signal based on the formation of complementary partners (HiBiT system). However, this signal-off approach is impacted by compound toxicity and requires molecular engineering, which can be challenging when using different cell types and primary cells. By contrast, no-wash immunoassays like HTRF (Homogeneous Time-Resolved Fluorescence) and Alpha (Amplified Luminescent Proximity Homogeneous Assay) can be used to monitor the impact of degraders on a POI over time, in their cellular context, and without the need for molecular engineering (Figure 3).
AlphaLISA SureFire Ultra and HTRF cell-based total protein assays are designed to quantify the expression level of a protein in a cellular lysate. The intensity of HTRF and Alpha signal is directly proportional to the concentration of the protein present in the sample.
AlphaLISA SureFire Ultra and HTRF cell-based total protein represent a straightforward technological platform which facilitates a comprehensive assessment of key degraders’ features such as degradation efficiency and selectivity towards a family of sibling proteins (eg BRD4, BRD2, BRD3) as well as mechanisms of action such as cytotoxicity, proteasome mediated degradation, and downstream signaling events. These assays quantify the expression level of endogenous proteins and are compatible with various biological models, from recombinant cell lines to more physiologically relevant samples such as tissues extracts. Therefore, combining HTRF or AlphaLISA Surefire Ultra cell-based total protein detection kits with HiBit engineered cell lines offers a deeper understanding of degraders’ mechanism of action. For instance, downstream signaling events can be easily monitored with HTRF or Alpha Total Kits used with HiBit recombinant cells. HTRF and Alpha also enables the assessment of biological response consistency between parental unmodified and HiBit cell lines. Identification of hit compounds obtained with HiBit technology can be easily confirmed with an orthogonal no-wash immunoassay method.
Another consideration is whether the selected degrader can pass through the cell membrane to find its intended POI. CETSA can provide the required information in a fast and easy way across different cell types without any modifications.
Complementary to target based screening methods, phenotypic-based screening approaches can be used to study the morphological changes induced by PROTACs. In this context, cell painting is a powerful high-throughput high-content screening and image analysis method to phenotypically characterize the response of a cell to a perturbagen (compound, drug, or gene). This approach provides critical phenotypic information and can be used to better understand PROTACs’ safety profiles4.
Future Outlook for TPD
The past few years have seen the field of TPD move from proof of concept to the development of degrader drugs that are now advancing in clinical trials. As our understanding of proximity-induced degradation grows and trial data are presented, it is predicted that investments will continue to sour, and breakthroughs will likely follow. Fundamental to the success of degraders is our continued ability to interrogate every step of the degradation pathway. With the right tools, technologies, and collaborative efforts, there is no doubt that degraders will soon realize their full potential and move closer toward safe and effective treatments for patients.
References
- Surka C, Jin L, Mbong N, Lu CC, Jang IS, Rychak E, Mendy D, et al. CC-90009, a novel cereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells. Blood. 2021 Feb 4;137(5):661-677. doi: 10.1182/blood.2020008676. PMID: 33197925; PMCID: PMC8215192.
- Békés M, Langley DR, Crews CM. PROTAC targeted protein degraders: The past is prologue. Nature Reviews Drug Discovery. 2022;21(3):181–200.
- Yang N, Kong B, Zhu Z, Huang F, Zhang L, Lu T, et al. Recent advances in targeted protein degraders as potential therapeutic agents. Molecular Diversity. 2023; doi:10.1007/s11030-023-10606-w
- Trapotsi M-A, Mouchet E, Williams G, Monteverde T, Juhani K, Turkki R, et al. Cell morphological profiling enables high-throughput screening for proteolysis targeting Chimera (PROTAC) phenotypic signature. ACS Chemical Biology. 2022;17(7):1733–44. doi:10.1021/acschembio.2c00076