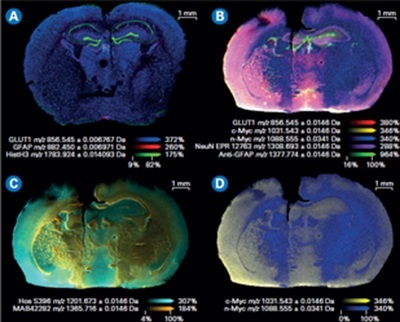
Composite color plots of extracted ion images of the mass tags used in a MALDI IHC workflow. A) Multiomics demonstration with lipids and drug target and the workflow on the same tissue shown here. B) Composite color plots of extracted ion images of additional multiplexed antibody tags from the MALDI IHC workflow. C) and D) Composite ion images using select mass tags to specifically compare targeted proteins of interest for this system.
by Kate Stumpo, PhD, Senior Market Manager, Bruker Scientific, LLC for Americas
Across many active areas of clinical research, one technique has established itself as key to unlocking the intricacies of molecular activity within the context of native tissue: matrix-assisted laser desorption ionization (MALDI) Imaging. This powerful methodology enables researchers to explore the biological mechanisms that regulate cellular interactions and maintain tissue homeostasis, to better understand, and, ultimately, inform our knowledge of the progression and treatment of disease.
MALDI imaging generates spatial distribution maps of metabolites, lipids, glycans, and peptides without the need for molecular markers or labeling; and larger proteins can be analyzed using appropriate markers.
Now, the latest developments encourage deeper analysis and visualization of molecular distributions within thin tissue sections and, for the first time, enable rapid multiplexed, multimodal, and multiomic analysis of a whole slide.
Cutting through the complexity
Since its inception in the early 1990s, MALDI Imaging has steadily grown in prominence, with each technical or informatics advance expanding its use in clinical research. The breakthrough in 2005[1] that enabled the accurate co-location of MALDI images with other non-mass spectrometry imaging data, cemented its status as a vital tool in cancer biology, neurobiology, toxicology and pharmacokinetics, for example.
At the heart of MALDI Imaging lies its ability to measure and visualize the spatial distribution of both endogenous and exogenous molecules within their native tissue context. While the concept of measuring the spatial distribution of molecules on tissue sections is not new[2], traditional methods such as immunohistochemistry (IHC) and in situ hybridization (ISH) have faced many limitations, particularly in scaling up to multiplexed analyses due to challenges associated with using numerous dyes or labels.
Today, MALDI Imaging methods can incorporate IHC capabilities for targeted protein imaging, facilitating rapid multiplexed, multimodal and multiomic analyses of entire slides. It allows untargeted, label-free analysis of metabolites, lipids, glycans and peptides, while enabling targeted analysis of larger protein molecules using appropriate labeling strategies. Typically applied to fixed fresh-frozen or formalin-fixed paraffin-embedded (FFPE) tissue sections, MALDI Imaging generates extensive datasets comprising thousands to tens of thousands of spectra, each representing intensities measured across numerous positions and describing hundreds of different molecules.
Integrating MALDI with IHC
Leaders in the field are looking for techniques that are able to to perform multiomics in one run, though to accomplish this several challenges needed to be overcome. The ability to add more markers and increase plex, along with higher resolution without any loss of sensitivity, perhaps being the core advancements required. In addition, more sophisticated informatics would be needed to process and analyze the much larger datasets generated by this approach. Solving these challenges has allowed researchers to combine the principles of IHC with the high sensitivity of MALDI, to achieve spatially resolved protein profiling within tissue samples, and enable the simultaneous identification and localization of multiple antigens with high resolution.
In practice, MALDI IHC uses novel photocleavable mass-tags linked to antibody probes to facilitate the development of high-throughput, multimodal, and multiomic MALDI workflows, capable of simultaneously visualizing both small molecules and intact proteins within the same tissue section.
Dual-labeled antibody probes enable the combination of multimodal mass spectrometry (MS) and fluorescent imaging for precise examination of specific intact proteins. The latest integrated workflows permit the simultaneous analysis of MALDI Imaging with IHC (Figure 1), permitting the concurrent analysis of more than a hundred distinct biomarkers in a single scan and eliminating the need for repetitive cycling.
Each probe carries a unique mass which generates highly multiplexed images. These images can be seamlessly integrated with other imaging techniques for multimodal data analysis, or subsequent MALDI Imaging sessions (Figure 2). This capability also permits the spatial mapping of metabolic pathways associated with specific proteins of interest and the quantitative evaluation of protein activity. As a result, researchers can precisely co-localize small-molecule drugs and their corresponding targets, including associated biomolecules involved in the cellular response to these drugs.
Studies using MALDI IHC workflows have shown the ability to provide high-plex, multiomic, and multimodal imaging of tissues.[3] Specifically, this integrated technology provides a platform to image 100+ protein biomarkers on a whole slide in around 40 minutes, at a resolution of 5 µm. Fluorescence microscopy, histopathological staining, and MALDI Imaging are integrated on one tissue section with no conjugation or metal tags required (Figure 3).
Uncovering novel biomarkers
Recent studies have showcased the capabilities of MALDI IHC to assist in finding biomarkers linked to disease, offering unprecedented opportunities in tissue pathology, early detection, prognosis, and precision medicine. This includes the successful demonstration of up to 121-plex MALDI-IHC on diverse tissue specimens such as mouse brain, human tonsil, and breast cancer tissues. Furthermore, the integration of novel techniques such as dual-labeled fluorescent photocleavable mass-tags (PC-MTs) antibodies and label-free small-molecule MALDI Imaging, has significantly extended the scope and effectiveness of the MALDI IHC approach.[4]
Insights into cancer and neurobiology
Beyond its applications in disease-based research, MALDI IHC is revolutionizing our understanding of complex biological processes, particularly in cancer biology and neurobiology. By elucidating the intricate relationships between cancer cells and their microenvironment, MALDI IHC serves as a powerful tool for unraveling the heterogenous cellular composition and offers valuable insight into tumor growth, invasion, and response to therapy. This comprehensive analysis allows researchers to identify potential biomarkers indicative of disease progression. Such investigations may, ultimately, allow tailored therapies for individual patients.
In neurobiology, MALDI Imaging provides a unique window into the distribution of neurotransmitters, neuropeptides, and associated proteins within the brain, shedding light on the molecular underpinnings of neurological diseases such as Alzheimer's disease, Parkinson's disease, and other psychiatric disorders. By mapping the distribution of these molecules, researchers can better understand the complex mechanisms underlying healthy brain function and pathological processes, potentially opening up new avenues for therapeutic interventions and drug development.
What comes next?
Recent developments in MALDI imaging – specifically the introduction of MALDI IHC – further cement the methodology at the heart of clinical research in many important disease areas. As a new high-performance, integrated multiplexed, multimodal and multiomic technique, researchers can now access intuitive workflows and generate a more comprehensive, high-quality dataset in order to better understand complex biological processes. With its ability to unravel the complexities of biological systems at the molecular level, MALDI IHC is poised to influence the future of disease-based research as the insights gained become targets for translational clinical research.
About the author: Kate Stumpo received her PhD in Analytical Chemistry from Texas A&M University in 2008. She worked in academia for 11 years before joining Bruker in 2021. After gaining experience as an Applications Scientist and Business Development Manager, she currently holds a Senior Market Manager position.
References
[2] Eisenstein, M. (2022). Seven technologies to watch in 2022. Nature, 601, 658–661.
[3] Lim, M. J., Yagnik, G., Henkel, C., Frost, S. F., Bien, T., & Rothschild, K. J. (2023). MALDI HiPLEX-IHC: Multiomic and multimodal imaging of targeted intact proteins in tissues. Frontiers in Chemistry, 11, 1182404. https://doi.org/10.3389/fchem.2023.1182404
[4] Yagnik, G., Liu, Z., Rothschild, K. J., & Lim, M. J. (2021). Highly Multiplexed Immunohistochemical MALDI-MS Imaging of Biomarkers in Tissues. Journal of the American Society for Mass Spectrometry, 32(4), 977–988. https://doi.org/10.1021/jasms.0c00473