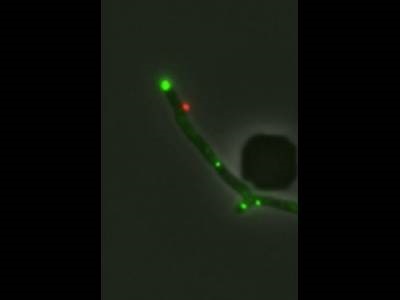
Infection of Mycobacterium smegmatis by a genetically engineered mutant of phage Fionnbharth. A single phage particle binds to the bacterial cell, seen as a red dot. Credit: Charles Dulberger
Bacteriophages – viruses that can infect and kill bacteria – present an opportunity to treat diseases for which other antibiotic treatments have become ineffective due to resistance. Bacteria can also develop resistance to phages, and the mechanisms of this resistance are not well understood. A team including researchers from Harvard University, MIT and the University of Pittsburgh have now used innovative live-cell fluorescence microscopy techniques to view the interaction between phages and bacteria in unprecedented detail, revealing the role of certain genes and proteins in infection and resistance, and offering insights for the future development of phage-based treatments.
The study involved the infection of Mycobacterium smegmatis with a mycobacteriophage known as Fionnbharth. First, the researchers utilized phages stained with red fluorophores to infect bacteria containing a probe known as N-QTF, which reacts with Ag85 mycolyltransferase to label sites of cell wall synthesis in the bacteria. The red fluorophore only activates when a cell membrane is compromised, allowing the binding sites of the phages on the bacteria to be identified. Time-lapse microscopy using a widefield fluorescence microscope showed that the phages preferred to attach to cell wall synthesis sites near the center and ends of the bacteria. The team then studied the impact of gene mutations on phage infection utilizing phages labeled with the red fluorophores and short DNA sequences known as MalO operators. When the phage infects M. smegmatis expressing Mal1 proteins linked with green fluorophores, the proteins bind to the MalO operators, allowing the spread of the phage to be further visualized through the green fluorescent signal.
The team performed experiments using bacteria with a deletion of the lsr2 gene, which encodes a protein that aids the bacteria in replicating its own DNA. The researchers found that deletion of lsr2 caused the M. smegmatis to become resistant to Fionnbharth. By labeling the Lsr2 protein, it was possible to observe the protein’s role in Fionnbharth infection, and time-lapse microscopy revealed that the protein moved to phage replication sites after infection. These findings suggest that the phage is able to hijack Lsr2 to replicate its own DNA and uncovers how mutations such as lsr2 resistance could enable bacteria to become phage-resistant. This research was published in Nature Microbiology.
“This paper focuses on just one bacterial protein. There are lots of different phages and lots of other proteins,” said co-corresponding author Graham F. Hatfull, noting the wide implications of the research and the techniques used to observe phage activity.
Using similar techniques to probe the interactions between different phages and bacteria could help scientists engineer phages that can more effectively avoid resistance. For example, mycobacteriophages like Fionnbharth could potentially be used to treat Mycobacterium tuberculosis and Mycobacterium abscessus, relatives of M. smegmatis that have become more difficult to treat due to antibiotic resistance.