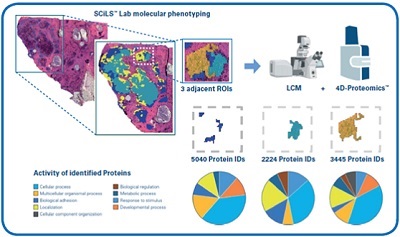
Figure 1: Illustration of MALDI Guided SpatialOMx analysis of breast tumor biopsy. Pathologist-annotated tumor regions are further segmented by SCiLS Lab based on molecular phenotyping derived from MALDI based lipid imaging. A very narrow region of tumor segmented into three molecular sub-regions is targeted for removal by LCM into collections of ~2000 cells from each sub-region. PASEF enables high sensitivity 4D-Proteomics from 160 ng of peptides injected into the timsTOF fleX to characterize the proteomes of each of the cellular subpopulations distinguished by GO annotation.
by Andreas Sonnen, Assistant Professor at University Medical Center Utrecht, and Michael L. Easterling, Imaging Business Director, Bruker Daltonics
Cancer is the second leading cause of death worldwide, with 10 million people dying from it every year.1 Tools that facilitate accurate cancer diagnosis and support potential treatment, especially in the early stages of disease, can direct clinicians to more effective treatment procedures. The heterogeneity of tumors presents a significant challenge in developing effective personalized treatments. Tumor heterogeneity refers to the differences between tumor cells of one patient or in the same type of tumor among multiple patients, and these differences can involve genes and/or proteins within the tumor.2 It means that not all tumor cells behave in the same way with regard to treatment response but also in reaction to analytical tools, for example immunohistochemistry. Over time, cancers can become more heterogeneous and resistant, which makes the treatment process increasingly difficult, thus spatial imaging of whole tumor slides, primary tumor and metastases, becomes progressively important.3
Unmet need-based oncology research
Personalized medicine is an emerging approach, especially in oncology departments, for tumor treatment and identification of biomarkers that advance prevention, diagnosis, prognosis, and therapeutics. Moving away from a one-size-fits-all- approach could improve overall patient care and ultimately and ideally result in a tailored treatment plan. Continued technological advances in Matrix-assisted laser desorption/ionization (MALDI) mass spectrometry (MS) hold great promise for developing a deeper understanding of the complexity of tissues and diseases.
Researchers need to gain a greater molecular and functional understanding of the cells comprising different cancers and their pre-existent tissue environment in order to improve future treatment, for example the immune cell environment accompanying many cancers. Personalized medicine has the potential to tailor therapy towards the oncogenic drivers of a specific tumor and its surrounding environment. In an attempt to obtain a new and more effective diagnosis, researchers are assigning tumors by molecular characteristics since they realize generic treatments such as chemotherapy and radiation are oversimplified, less effective and can cause unnecessary side effects. In multi-omics the combination of several omics datasets from different molecular levels, e.g. DNA, RNA, proteins, lipids, glycans and metabolites, can identify patient subgroups on the basis of biological features of the underlying diseases.4 Adopting this approach can provide a more comprehensive overview. In this way useful biomarkers of disease processes can be uncovered potentially indicating targeted treatment options for patients.
Supporting personalized medicine
Researchers at the University Medical Center Utrecht (UMC Utrecht) use MALDI-MS imaging to improve clinical diagnosis and pathology research. The UMC Utrecht has over 11,000 employees and is one of the largest public healthcare centers in the Netherlands. Its current research is centered around improving patient care from diagnosis to therapeutics, focusing on discovering new treatments for disease. MALDI is a ‘soft’ ionization technique that provides high responses for the unfragmented molecular ion and can be applied to a broad variety of molecules ranging from small drug-like compounds up to larger peptides and intact proteins.5 Due to recent technological innovations molecular profiles from thousands of molecules at each image pixel without the breakdown of tissue morphology can be achieved at nearly cellular resolution with fast scanning times. This enables researchers to assess molecular tumor heterogeneity combined with classical histology of the same tissue section.6
When looking at the proteome, lipidome or metabolome, the spatial distribution of compounds contains valuable information. MALDI-MS imaging is a powerful tool that enables their visualization.
Exploring MALDI-TIMS TOF MS
In MALDI-TOF MS, the samples are accelerated into a TOF analyzer, providing access to the mass spectra of molecules such as proteins, peptides, lipids, carbohydrates, and nucleotides.7 MALDI-TOF MS is advantageous because of the low requirements on sample preparation. The researchers in the UMC Utrecht pathology department are combing MALDI-TOF MS with trapped ion mobility separation (TIMS) with the aim of achieving better clinical diagnosis, because it can separate isobaric and isomeric metabolites, lipids, peptides and glycans found in complex samples. This application of TIMS within the MALDI workflow allows for the separation of molecules depending on the shape of the ions. TIMS is based on the collisional cross section (CCS) of detected molecules and allows improvement when measuring the validity of an analyte, which supports researchers in meeting expected quality criteria by providing extra detail. CCS-enabled software matches with spatial MALDI-TIMS imaging data alongside omics results, making a perfect pairing for vital morphological context in identification lists.
The UMC Utrecht is able to expand its medical research capabilities through advanced technology such as TIMS because it can differentiate isomeric distributions where mass resolution fails, to reveal the true spatial localization of an analyte. The more understanding of complex samples that is gained, the closer the team is to improving patient outcomes. A study conducted by UMC Utrecht used laser-capture microdissection (LCM) to extract specific tissue regions to gain further insight into cancer treatment. The extractions can be used with MALDI-TOF MS to get spectral information to create a molecular map of the tumor including tumor heterogeneity. This molecular information could help physicians in deciding on the treatment best suited for the patient. Identifying for example modifications in protein expression or the metabolic state of the cell could uncover new targets or molecular markers for heterogenous tumors and, again, could help researchers develop other treatment possibilities and make personalized medicine more accessible.
Moving toward the future
When looking at the scientific and social challenges being faced at hospitals and medical centers, there seems to be a shifting focus to continue the push for personalized medicine to be more easily accessible. At the UMC Utrecht, the team has two primary goals: one, to understand heterogeneity of diseases using MALDI-MS and, based on this, to find new therapeutic and prognostic biomarkers for different diseases using MALDI-MS.
Although advances in diagnosis speed and accuracy have propelled the field in recent years, MALDI-MS technologies are set to transform our knowledge of molecular mechanisms and drive pathology research. Thanks to its ability for rapid and sensitive profiling of a large number of molecules simultaneously, MALDI-MS could lead to new putative disease markers in cancer. The innovative developments undertaken by the team at UMC Utrecht place them in a pivotal position in supporting personalized medicine development and how they can make the shift from a laboratory to a clinical setting. MALDI-MS imaging currently occupies the field of cancer research and, in combination with TIMS, looks optimistic for new discoveries to aid patient care while keeping costs at a minimum.
References
1. World Cancer Day, What is cancer?,https://www.worldcancerday.org/what-cancer
2. National Cancer Institute, Tumor Heterogeneity. https://www.cancer.gov/publications/dictionaries/cancer-terms/def/tumor-heterogeneity
3. Dagogo-Jack, I., Shaw, A. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 15, 81–94 (2018). https://doi.org/10.1038/nrclinonc.2017.166
4. Heo YJ, Hwa C, Lee GH, Park JM, An JY. Integrative Multi-Omics Approaches in Cancer Research: From Biological Networks to Clinical Subtypes. Mol Cells. 2021 Jul 31;44(7):433-443. doi: 10.14348/molcells.2021.0042. PMID: 34238766; PMCID: PMC8334347.
5. M.W. Duncan, D. Nedelkov, R. Walsh and S.J. Hattan, Applications of MALDI mass spectrometry in clinical chemistry, Clinical Chemistry, 2016, 62: 134–143, https://doi.org/10.1373/clinchem.2015.239491.
6. Flach RN., Fransen NL., Sonnen AFP., Nguyen TQ., Breimer GE., Veta M., Stathonikos N., van Dooijeweert C., van Diest PJ. Implementation of Artificial Intelligence in Diagnostic Practice as a Next Step after Going Digital: The UMC Utrecht Perspective. Diagnostics (Basel). 2022 Apr 21;12(5):1042. doi: 10.3390/diagnostics12051042. PMID: 35626198; PMCID: PMC9140005.
7. Moskowitz, J. and Profile, F., 2022., MALDI-TOF MS: An evolving approach to cancer profiling., Today's Clinical Lab. https://www.clinicallab.com/trends/clinical-mass-spectrometry/maldi-tof-ms-an-evolving-approach-to-cancer-profiling-26470#:~:text=Matrix%2Dassisted%20laser%20desorption%20ionization,a%20wide%20range%20of%20cancers.