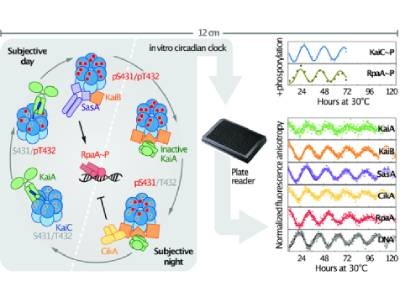
The biological clock, or circadian rhythm, that functions within our cells is what keeps our body in tune with the cycle of day and night; the same goes for the circadian clock of other organisms such as the single-celled cyanobacteria. This rhythmic cycle involves repeated interactions between proteins and DNA, which can be difficult to monitor in real time and in complete detail within an intact organism. Researchers at UC Santa Cruz, UC Merced and UC San Diego campuses recently assembled a functioning biological clock completely in vitro, allowing for detailed study of its components and their interactions using fluorescent labeling.
The UC teams built on previous work by Japanese researchers, who built a cyanobacterial circadian oscillator in 2005. This oscillator consisted of the three proteins KaiA, KaiB and KaiC, which perform the circadian clock’s basic 24-hour timekeeping. In a full cyanobacterial biological clock, these oscillator proteins transmit signals through other proteins to control gene expression throughout the cycle. The UC researchers added additional components to the basic oscillator, including the two kinase proteins SasA and CikA, along with the DNA-binding protein RpaA and its DNA target.
The final clock functions autonomously as SasA and CikA respectively activate and deactivate the RpaA in order to rhythmically bind and unbind the DNA, according to UC Merced researcher and professor Andy LiWang. Within cyanobacteria, this binding and unbinding occurs at more than 100 different sites in the organism’s genome, regulating the expression of different genes important for the bacteria’s health and survival, said LiWang, who is a corresponding author on the paper.
Using fluorescent labeling, the researchers tracked how their clock’s components interacted with one another in real time as the system oscillated continuously for several days or even weeks. The research showed how SasA and CikA increased the robustness of the oscillator, which would ordinarily stop functioning when the Kai A, B and C proteins were working alone. The researchers performed in vivo tests and found that their in vitro clock was surprisingly consistent with how the cycle functioned in living bacteria. The team also explored the genes involved in circadian rhythm disruption in an arrhythmic strain of cyanobacteria; a single mutation in the gene for RpaA was identified that impacts the DNA binding efficiency of the protein. This research was published in Science.
“A single amino acid change in the transcription factor makes the cell lose the rhythm of gene expression, even though its clock is intact,” said coauthor Susan Golden, director of the Center for Circadian Biology at UC San Diego. “The real beauty of this project is how the team drawn from three UC campuses came together to pool approaches toward answering how a cell can tell time.”
This research could open the door for more advanced research of circadian rhythms through single-cell microscopy using artificial cells implanted with the in vitro system, and possibly allow circadian clocks to be engineered for further synthetic biology applications, the author wrote.
Photo: A diagram showing the cycle of interactions between the components in an in vitro biological clock reconstituted by UC scientists. Credit: Andy LiWang