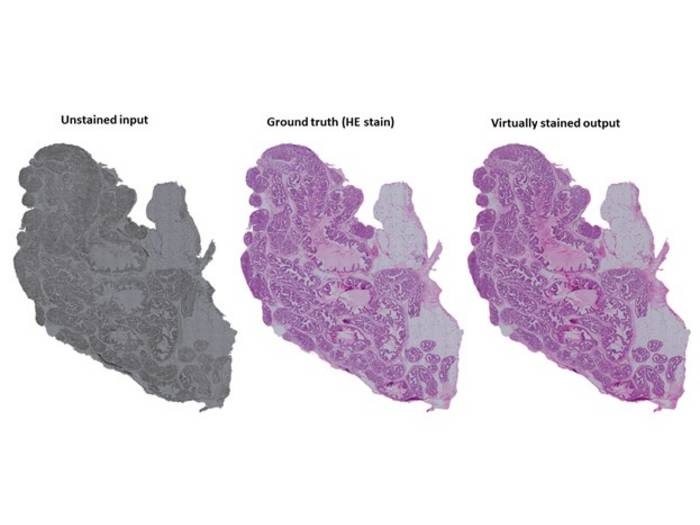
An example of virtual staining of tissue. Unstained tissue on the left, chemically stained tissue in the middle and virtually stained tissue on the right. The examples are prostate tissue. Credit: Pekka Ruusuvuori
Hematoxylin and eosin (HE) staining enables pathologists and researchers to clearly view the structures of tissues that would be nearly impossible to see without staining, even under a microscope. However, the staining process is very time- and labor-intensive, requires hazardous reagents, and is irreversible, preventing the same tissue sample from being used for other tests. A team of researchers from the University of Eastern Finland, University of Turku and Tampere University have developed an alternative to chemical staining that could overcome these challenges: a computational technique based on artificial intelligence that enables the “virtual staining” of unstained, imaged tissues.
The researchers trained unsupervised and supervised deep learning models – generative adversarial neural networks designed for image-to-image translation – on tens of thousands of “tiles” taken from brightfield microscope images of unstained and stained histological tissue sections. The reference images included a wide range of tissue types including epithelial tissue, muscle tissue, liver and testis. The team then tested accuracy of the AI models on samples that were imaged both before and after staining, allowing the virtually stained image to be compared to that produced through chemical staining. The researchers also further optimized their method by determining which sample preparation techniques and network parameters produced the best virtually stained images.
The team found that both models produced virtually stained images that greatly resembled those produced through traditional chemical staining, although accuracy was improved when using the supervised model. Deparaffanized tissue sections were virtually stained more accurately than unprocessed paraffin-embedded samples, although the authors noted that improvements in algorithms could be made to overcome this problem, which would better enable sections to be reused for other analyses such as Fourier transform infrared (FTIR) and Raman spectroscopy. Preparing thinner sections, between 3 and 8 μm, which are commonly used in histopathology workflows, resulted in better virtual staining results than using thicker sections between 12 and 20 μm. Results were also found to be more favorable when tissues were imaged at 20x magnification compared with 40x magnification.
Optimization of the supervised generative adversarial neural network resulted in greater structural similarity scores, signal-to-noise ratio and nuclei count accuracy when simple convolutions were replaced with dense convolution units, the authors wrote. The virtual staining method was first described in Laboratory Investigation in January, while further model optimization was reported in a paper published in Patterns this month.
“Deep neural networks are capable of performing at a level we were not able to imagine a while ago,” said Umair Khan, first author of the most recent paper and doctoral researcher at the University of Turku. “Artificial intelligence-based virtual staining can have a major impact toward more efficient sample processing in histopathology.”
The virtual staining method was developed using the high-performing computing services at CSC - IT Center for Science. The developers noted that virtual staining could become a feasible solution for any laboratory with a conventional brightfield microscope and suitable computer system. Removing the need to chemically stain histological tissue samples could result in immense time and resource savings, reduce the use of toxic chemicals and reduce the need to acquire multiple tissue samples, as the same sample could be reused for other testing methods.